UNIT 3 - Digital Epidemiology Tool: PATHOGENWATCH
 OUTLOOK
OUTLOOK
At the end of this unit, the participants shall be able to:
identify the key features of Pathogenwatch; and
recognize the analyses that can be generated through Pathogenwatch
 LESSON PROPER
LESSON PROPER
Pathogenwatch is a free web based tool for processing and visualization of microbial genome sequences in phylogenetic and geographical contexts. First, this genomic tool allows you to upload microbial genomes to access an array of automatic analyses which includes the following:
Species assignment using “Speciator”.
Over 20 core genome MLST schemes.
Typing with classic MLST, Genotyphi, NG-MAST, PopPUNK and others.
Antibiotic resistance prediction.
Interrogate the database using cgMLST-based clustering and SNP trees.
Moreover, this tool provides extensive collections of publicly available microbial sequences, gathered by publication or survey where possible. The imported genomes are automatically annotated within the in-house pipelines of Centre for Genomic Pathogen Surveillance (CGPS) and the results available for download. Pathogenwatch also utilizes a clustering-based system allows the identification of near genomic neighbors for both local and global context mapping of your uploaded genomes. Lastly, this genomic tool makes use of powerful selection tool to integrate public genomes with your own and create a single collection. This collection can be viewed by public to facilitate sharing of insights among researchers.
| Uploading Genomes to Pathogenwatch | |
|---|---|
Open Pathogenwatch Navigate to https://pathogen.watch/upload and click on the “Single-genome FASTAs” image in the center of the screen. |
 |
Upload your file Click the plus button in the bottom right corner and then navigate to where your FASTA files, or FASTQ files are stored. |
 |
| View the genomes |  |
| Create and view collection | 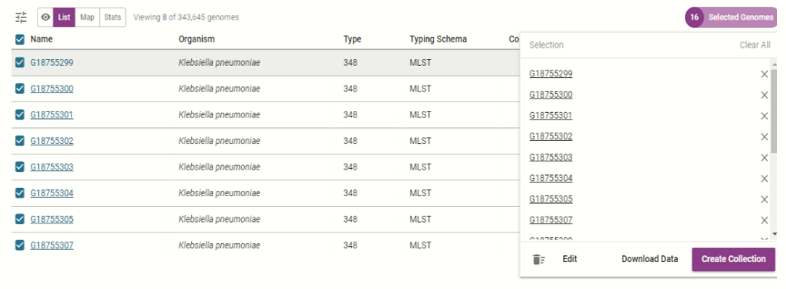 |
| Exploring Genomes in Pathogenwatch |
|---|
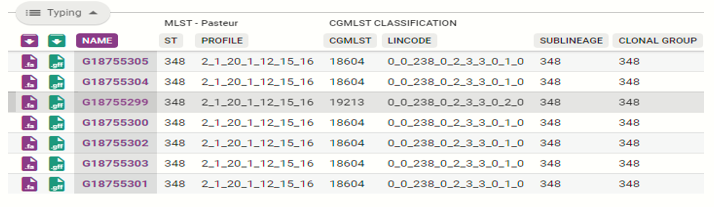
At the top left of the table you’ll see a lozenge button that says “Typing.” This will provide information on the multi locus sequencing type of the organism following the Pasteur scheme.
|
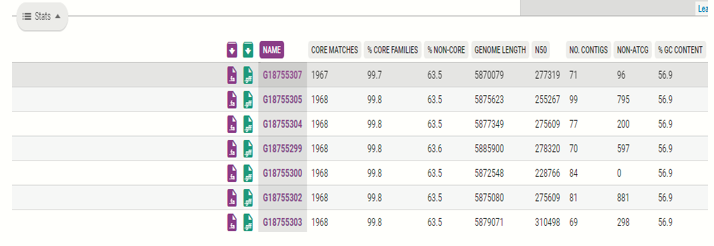
Next, click on the “Stats”, to show the quality control metrics for each of the isolate analyzed.
|
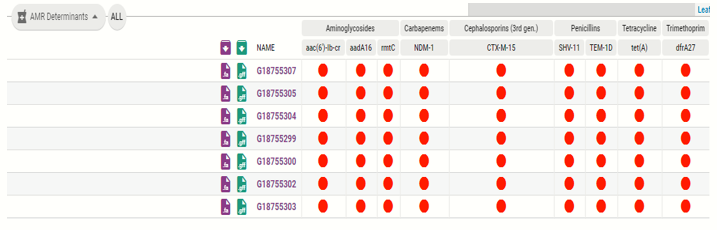
Then, click on “AMR Determinants” to view the AMR genes observed in the isolates.
|
In addition to providing big picture information about how isolates relate to each other, Pathogenwatch also provides individual reports that can be downloaded or printed to share with partners or submitters. The figures below show the generated report on one isolate.


 BRAINWORK
BRAINWORK
To learn and explore more on the advanced features of Pathogenwatch, click on the link below:
 FLICK
FLICK
For your further reading, the following journal articles below utilized Pathogenwatch for their study analysis.
| Study Title | Author/s | DOI |
| Rapid Genomic Characterization and Global Surveillance of Klebsiella Using Pathogenwatch | Argimon et al. 2021 | https://doi.org/10.1093/cid/ciab784 |
| A community-driven resource for genomic epidemiology and antimicrobial resistance prediction of Neisseria gonorrhoeae at Pathogenwatch | Sanchez-Buso et al. 2021 | doi: 10.1186/s13073-021-00858-2 |
| A global resource for genomic predictions of antimicrobial resistance and surveillance of Salmonella Typhi at pathogenwatch | Argimon et al. 2021 | doi: 10.1038/s41467-021-23091-2 |